Targeting Disease with Precision: A Primer on Antibody Discovery

The earliest reference to the potential of antibodies for treatment in humans came from an 1890 publication, authored by Dr. Shibasaburo Kitasato and Dr. Emil von Behring, describing how serum from animals with diphtheria and tetanus could cure those diseases in other animals.
Since then, significant milestones have been achieved at a continuous pace. The chemical structure of antibodies was published in 1959, the first atomic resolution structure of an antibody fragment in 1973, and a technique to create identical antibodies, or monoclonal antibodies, emerged in 1975 to catalyze the field of modern antibody research and discovery.
The first therapeutic monoclonal antibody, Muromonab-CD3, was approved by the FDA in 1986 with the second, which incidentally was the first approved-for-use chimeric antibody, following 8 years later. The milestone 50th approval came 21 years later, crossed a hundred in 2021, and according to recent data, an estimated 175 antibody therapeutics are in regulatory review or approved with an additional 1200 currently in clinical studies.
By 2027, monoclonal antibodies are projected to drive biologics sales revenue past that of small molecules. As we head to that seminal milestone, here’s a quick back-to-basics overview of some of the key terms and concepts underlying antibody drug discovery.
Antibodies
What are antibodies?
Antibodies, or Immunoglobulins (Ig), are specialized proteins that are part of the adaptive immune system and help recognize, neutralize, and eliminate disease-causing pathogens. They are a key component of the natural immune response to infection.
How do antibodies work?
Antibodies recognize and bind to specific proteins on the surfaces of antigens such as viruses, bacteria, etc. When exposed to a foreign substance for the first time, the body’s immune cells produce antibodies that target specific proteins associated with that particular antigen, for example, the unique spike protein SARS-CoV-2. Even after the threat of infection has been neutralized, these antibodies, along with memory cells of antibody-producing immune cells, remain in the body to provide a faster immune response when exposed to the same pathogen in the future. This humoral immunological memory is created by long-lived plasma cells (LLPCs), which produce the protective antibodies, and memory B cells, which are quiescent cells that remember past exposure to enable a quick response.
Structure of antibodies
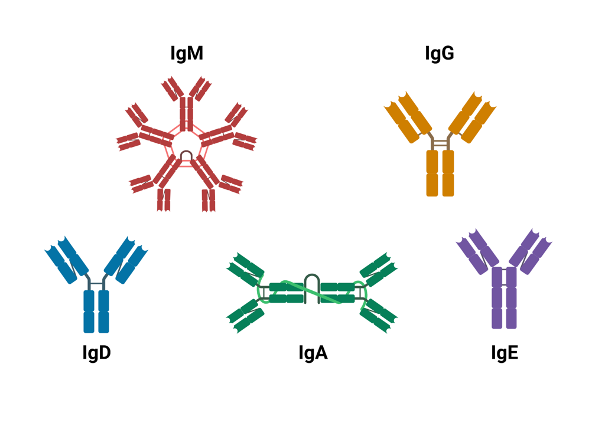
SOURCE: Wikimedia Commons
All antibodies share the same Y-shaped structure consisting of two identical light chains (LCs) and two identical heavy chains (HCs). Most natural systems feature one LC paired with one HC associated with another identical heterodimer to create the final form. In mammals, LC can be either of two functionally similar types, lambda (λ) and kappa (κ) with each type consisting of two domains, a constant domain (CL) and a variable domain (VL). HCs, on the other hand, can be any of five isotypes, IgA, IgD, IgE, IgG, and IgM. The number of constant (C) and variable (V) domains varies across these 5 types with IgAs, IgDs, and IgGs having three constant and one variable domains and IgEs and IgMs having one variable and four constant domains. Each of these variations differs slightly in terms of shape — for instance, IgG, is a straightforward Y compared to IgM which looks like five Ys stacked together — as well as function within the adaptive immune system. For instance, IgM is the primary antibody response to pathogens and its secreted form enables strong binding to the pathogen, IgG, the most abundant antibody, neutralizes toxins and opsonizes bacteria, and IgA is the first defense for mucosal membranes. A clear understanding of the relationships between the structure and functions of antibodies is vital to the successful development of antibody-related therapeutics.
Monoclonal antibodies
What are monoclonal antibodies?
Monoclonal antibodies (mAbs) are man-made proteins designed to simulate the immune system’s natural ability to make specialized proteins, or antibodies, that can recognize and destroy target antigens. They are called monoclonal because antibodies created in the laboratory are clones, i.e. exact copies, of just one antibody and bind only to one antigen. Polyclonal antibodies (PAbs), on the other hand, are a collection of antibodies, secreted by different B cell lineages in response to an antigen, that can recognize and react to several different isotopes of the same antigen.
How are mAbs produced?
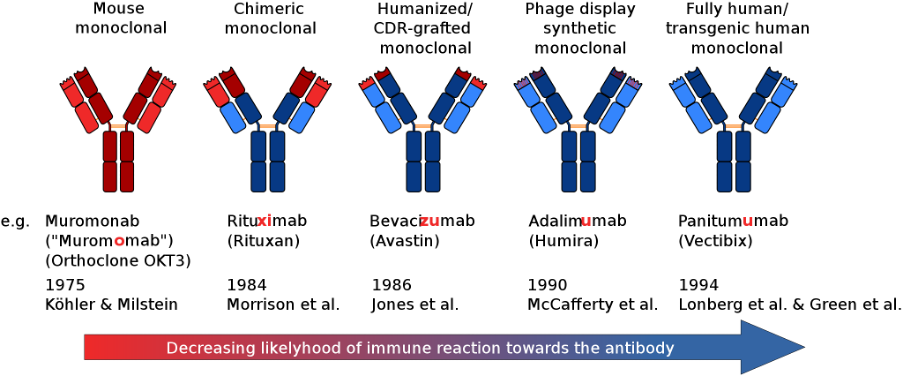
SOURCE: Wikimedia Commons
There are 4 different ways to make mAbs in the laboratory.
Murine (The names of treatments developed using this approach end in -omab): Early monoclonal antibodies were created with mouse myeloma cells. Even though these products were invaluable laboratory research, the potential of these murine-derived antibodies was limited by human anti-mouse antibody responses. Consequently, the focus turned to antibody engineering techniques that could lower the risk of immune reactions.
Chimeric (names end in -ximab): Chimeric antibodies were the first engineered improvement and involved replacing murine constant regions with human constant regions. This approach, of sequentially replacing mouse sequence-derived amino acids, was able to significantly reduce the immunogenicity of murine antibodies. Chimeric antibodies are molecules containing domains from different species, such as chimeric equine-human and canine-derived chimeric mAbs.
Humanized (names end in -zumab): Humanization was the next evolutionary leap in the development of animal-derived antibodies and the first product appeared in 1997 with the FDA approval of Daclizumab for the prevention of organ transplant rejection. Approximately 40% of FDA-approved antibodies are humanized. Humanized antibodies, mAbs that are mostly, but less than fully, human, are produced using different techniques. They could be nonhuman antibodies where some proportion of the animal sequences have been replaced with human sequences using techniques like complementary-determining regions (CDRs), grafting, resurfacing, and hyperchimerization. It could even be fully human mAbs that have been engineered to be less than fully human or to acquire sequences beyond the human repertoire.
Fully Human (names end in -umab): Human sequence-derived antibodies, unlike humanized antibodies, do not contain murine-sequence-derived CDR regions. There are currently three platforms available for the development of human mAbs: phage display technologies, transgenic mice, and yeast display. At last count, there were at least 28 FDA-approved human mAbs of which a majority (19) were derived from transgenic mice and the rest with phage-display technology. In recent years, yeast surface displays (YSDs) have emerged as a robust methodology for the isolation of mAbs against various antigens, though they are yet to deliver a product to market, However, phage display, the technique that delivered the first fully human therapeutic antibody, adalimumab, in 2002, continues to be the gold standard for isolating mAbs, especially for difficult-to-screen targets.
How do mAbs work?
Monoclonal antibodies today have a wide range of clinical and experimental applications. Initially, mAbs in clinical medicine were designed for activity against specific immune cells, progressing to specific cytokines and on to inhibiting the activity of specific enzymes, cell surface transporters, or signaling molecules. As the use of mAbs continues to expand across diverse therapeutic applications, they are also being designed to work in different ways based on the protein being targeted.
In the case of cancer, for example, there are two broad ways mAbs can work as an immunotherapy. One, they can trigger the immune system by attaching themselves to otherwise hard-to-spot cancer cells in a process called antibody-dependent cell-mediated cytotoxicity (ADCC). Two, they could act on cells of the immune system to help attack cancer cells in a type of immunotherapy called checkpoint inhibitors. In fact, a particular monoclonal antibody drug may function by more than one means. Apart from flagging cancer cells and blocking inhibitors, mAbs could also work by triggering cancer cell-membrane destruction, blocking the connection to proteins that promote cell growth to prevent cancer growth and survival, and blocking protein-cell interactions to inhibit the development of new blood vessels that are required for a tumor to survive.
So, that was a quick dive into the basic concepts related to antibodies in this first article in our three-part series on antibody drug discovery and development. We’ll continue in the next with a closer look at the role of antibodies in modern drug development, examine some of the formats of approved and experimental antibodies, and explore the scope & potential in-silico antibody discovery.
Until then.
Subscribe to our Blog and get new articles right after publication into your inbox.
Subscribe to our blog:






